
Chemical Nature of Enzymes
Enzymes of Class 11
Chemical Nature of Enzymes
Most enzymes are proteins except few, but all proteins are not enzymes.
On the basis of chemical nature enzymes are of two types, simple and conjugate.
Simple Enzymes are entirely made of proteins only. Additional nonprotein substance or group is absent. The active sites reside on the protein moiety, e.g. pepsin, trypsin, lysozyme.
Conjugate Enzymes are enzymes which possess two parts - a large thermolabile protein part or apoenzyme and a small dialysable thermostable nonprotein part called cofactor.
The complete conjugate enzyme made of apoenzyme and cofactor is called holoenzyme.
Enzyme cofactors are non-protein components required for efficient activity of enzymes.
Cofactors may vary from simple inorganic ions to complex organic molecules, and may either remain unchanged at the end of a reaction or be regenerated by a later process. There are three recognised types of
cofactor : inorganic ions, prosthetic groups and coenzymes.
Inorganic ions (enzyme activators) : thought to mould either the enzyme or the substrate into a shape that allows an enzyme/substrate complex to be formed, hence increasing the chances of a reaction occurring between them and therefore increasing the rate of reaction catalysed by that particular enzyme. For example, salivary amylase activity is increased in the presence of chloride ions.
Prosthetic groups (for example FAD, haem) :
If the cofactor is tightly bound to the enzyme on a permanent basis it is known as a prosthetic group.
Prosthetic groups are organic molecules. They assist the catalytic function of their enzymes, as in flavine adenine dinucleotide (FAD).
FAD contains riboflavin (vitamin B 2 ), the function of which is to accept hydrogen. FAD is concerned with cell oxidation pathways and is part of the respiratory chain in respiration.
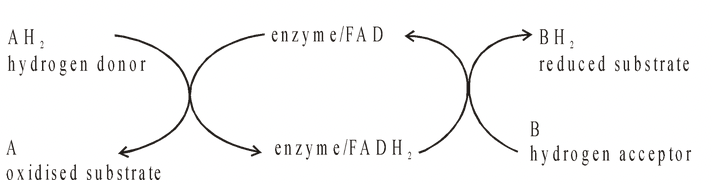
Net effect : 2H transferred from A to B. One enzyme acts as a link between A and B. Both AH2 and B fit into the active site and FAD passes H 2 from one to the other.
Haem is an iron-containing prosthetic group. It has the shape of a flat ring (a ‘porphyrin ring’ as is found in chlorophyll) with an iron atom at its centre.
Haem is the prosthetic group of cytochromes where it acts as an electron carrier. In accepting electrons the iron is reduced to Fe(II); in handing on electrons it is oxidised to Fe(III). It takes part in oxidation/reduction reactions by reversible changes in the valency of the iron.
Haemoglobin and myoglobin are oxygen-carrying proteins that contain haem groups. Here the iron remains in the reduced, Fe(II) form.
Haem is found in catalases and peroxidases, which catalyse the decomposition of hydrogen peroxide into water and oxygen. It is also found in a number of other enzymes.
Coenzymes (for example NAD, NADP, coenzyme A, ATP)
Like prosthetic groups, coenzymes are organic molecules which act as cofactors, but unlike prosthetic groups they do not remain attached to the enzyme between reactions.
All coenzymes are derived from vitamins. e.g. NAD (nicotinamide adenine dinucleotide) derived from the vitamin nicotinic acid (niacin) and can exist in both a reduced and an oxidised form. In the oxidised state it functions as a hydrogen acceptor.
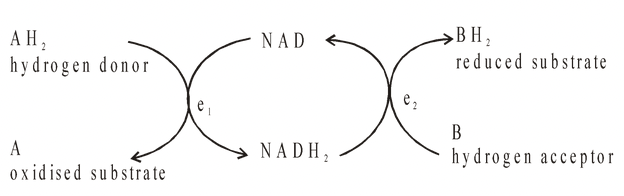
where e1 and e2 are two different dehydrogenase enzymes.
Net effect : 2H transferred from A to B. Here the coenzyme acts as a link between two different enzyme systems e1 and e2.
All enzyme names end in suffix - ase, except some old names, e.g. ptyalin, pepsin, trypsin. Some old names indicate the source but not the action, e.g. papain from Papaya, bromelain from Pineapple of family Bromeliaceae.
In modern system enzyme names are given after
(i) Substrate acted upon, e.g. sucrase (after sucrose), lipase, protease, nuclease, peptidase, maltase
(ii) Chemical reaction e.g. dehydrogenase, oxidase, carboxylase, decarboxylase etc.
Group names are often qualified by the addition of the name of substrate, e.g. succinic dehydrogenase, isocitric dehydrogenase, glutamate pyruvate transaminase, DNA polymerase.
DNA polymerase catalyses synthesis of DNA segments through polymerisation of deoxyribonucleotides.
Glutamate-pyruvate transaminase transfers amino group (–NH2) from glutamate to pyruvate.
In older times, enzymes were classified into two broad categories :
(i) Hydrolysing - Catalysing hydrolysis of larger molecules into smaller ones e.g. carbohydrases or amylases, proteases, lipases, esterases, phosphorylases, amidases. Digestive enzymes are hydrolysing in nature.
They are often grouped into three types - proteolytic, amylolytic and lipolytic.
(ii) Desmolysing - Catalysing reactions other than hydrolysis e.g. aldolases, dehydrogenases, oxidases, peroxidases, catalases, carboxylases etc.
The modern system of enzyme classification was introduced by International Union of Biochemistry (IUB) in 1961. It groups enzymes into the following six categories :
1. Oxidoreductases : They take part in oxidation and reduction reactions or transfer of electrons. Oxidoreductases are of three types - oxidases, dehydrogenases and reductases, e.g. cytochrome oxidase (oxidises cytochrome), succinate dehydrogenase, nitrate reductase.
2. Transferases : They transfer a group from one molecule to another e.g. glutamate pyruvate transaminase (transfers amino group from glutamate to pyruvate during synthesis of alanine). The chemical group transfer does not occur in the free state :
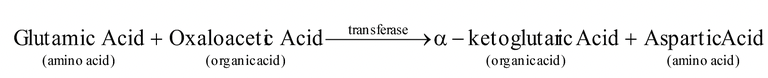
3. Hydrolases : They break up large molecules into smaller ones wiht the help of hydrogen and hydroxyl groups of water molecules. The phenomenon is called hydrolysis. Digestive enzymes belong to this
group, e.g. amylase (hydrolysis of starch), sucrase, lactase.
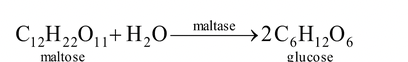
4. Lyases : The enzymes cause cleavage, removal of groups without hydrolysis, addition of groups to double bonds or reverse, e.g. histidine decarboxylase (breaks histidine to histamine and CO 2 ), aldolase (fructose-1, 6-diphosphate to dihydroxy acetone phosphate and glyceraldehyde phosphate).
Fructose1, 6-diphosphate
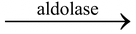 Dihydroxy acetone phosphate + Glyceraldehyde phosphate.
Dihydroxy acetone phosphate + Glyceraldehyde phosphate.
5. Isomerases : The enzymes cause rearrangement of molecular structure to effect isomeric changes. They are of three types
(i) Isomerases (aldose to ketose group or vice-versa)
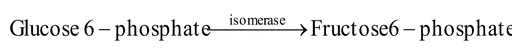
(ii) Epimerases (change in position of one constitutent or carbon group)
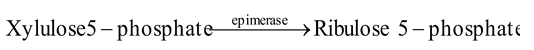
(iii) Mutases (shifting the position of side group).

6. Ligases (Synthetases) : The enzymes catalyse bonding of two chemicals with the help of energy obtained from ATP, e.g. pyruvate carboxylase. It combines pyruvic acid with CO2 to produce oxaloacetic acid.
Pyruvic acid + CO
2
+ ATP + H
2
O
 Oxaloacetic acid + ADP + Pi
Oxaloacetic acid + ADP + Pi





