
Genomic Library: A genomic library is a collection of DNA fragments that represent the entire genome of an organism. These DNA fragments are stored in identical vectors, with each vector containing a different piece of the genome. Scientists first extract DNA from the organism's cells and cut it into specific-sized fragments using restriction enzymes to create a genomic library. These fragments are then inserted into vectors using DNA ligase.
The vectors are introduced into a host organism, such as Escherichia coli or yeast, with each host cell carrying only one vector molecule. This allows for easy amplification and retrieval of specific DNA fragments from the library for further analysis. Genomic libraries are essential for sequencing applications and have been instrumental in sequencing the entire genomes of various organisms, including humans and several model organisms. The article below details the genomic library, its uses, steps of construction, and more.Also Check:
What is a Genome?
A genome is the full set of DNA in an organism, including all of its genes. It carries all the information needed to build and maintain that organism. In humans, the complete genome has over 3 billion DNA base pairs in every cell’s nucleus. This genome includes genes that code for proteins, genes that do not code for proteins and other types of DNA. Most eukaryotic organisms, including humans, have two copies of each chromosome, but when we talk about the genome, we mean just one copy of each chromosome.Genomic Library Definition
A genomic library is a collection of clones that include all the DNA from a particular organism. For example, an E. coli genomic library contains all the E. coli genes as well as the non-coding regions of the E. coli genome. This allows any desired gene to be retrieved and studied. To create a genomic library, total cell DNA is purified and partially digested with restriction enzymes to produce fragments. These fragments are then cloned into suitable vectors such as λ replacement vectors, cosmids, yeast artificial chromosomes (YACs), bacterial artificial chromosomes (BACs), or P1 vectors. Genomic libraries, consisting of colonies with DNA fragments, can be stored for many years and shared among research groups. For bacteria, yeast, and fungi, the number of clones required for a complete genomic library is manageable. However, for plants and animals, the number of clones needed is so large that finding a specific gene becomes very challenging. In these cases, a specialized library focusing on a specific cell type rather than the whole organism may be more practical.Also Check:
Genomic Library Diagram
The Genomic Library diagram is as follows:
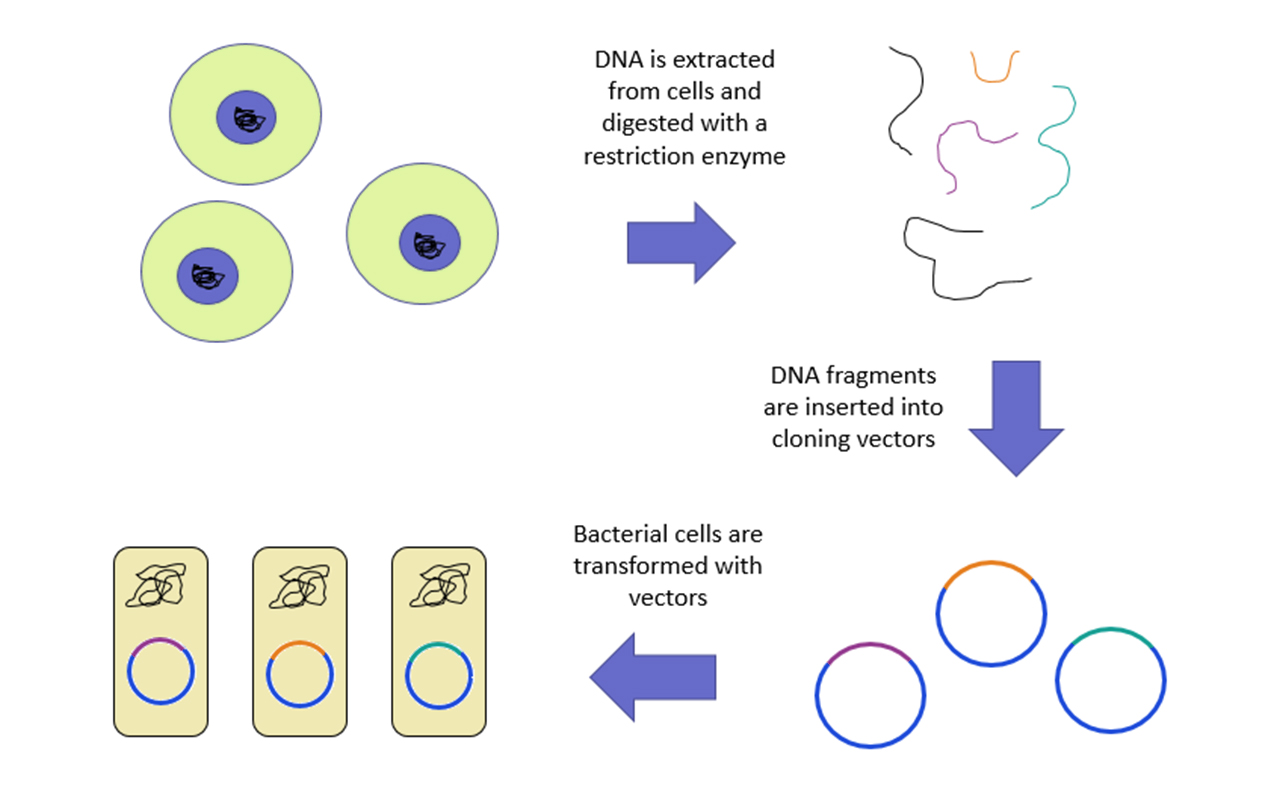
Genomic Library Construction
The first step in any DNA sequencing project is constructing a genomic library. A genomic library contains all the DNA sequences present in an organism's genome, including expressed and non-expressed genes, exons, introns, promoter and terminator regions, and intervening DNA sequences. An organism's genomic DNA is extracted and then cut into smaller fragments using a restriction enzyme to create this library. Numerous recombinant DNA molecules must be produced to cover the entire genome. Gene cloning involves five main steps:- Isolation of Genomic DNA and Vector
- DNA Fragment Generation
- Restriction Digestion
- Mechanical Shearing
- Ligation of DNA into a Vector System
- Transformation of Recombinant DNA into a Host Cell
- Library Screening
| NEET Biology Notes | |||
|---|---|---|---|
| Embryo | Funaria | Selaginella | Malvaceae |
| Polyembryony | Apomixis | Pinus | Pollen Grains |
| Chara | Volvox | Endosperm | Equisetum |
1. Isolation of Genomic DNA and Vector
Isolating genomic DNA is a straightforward technique that follows standard protocols or uses a ready-to-use DNA extraction kit. This process includes cell lysis, removal of proteins and impurities, DNA precipitation, DNA purification, and DNA elution. The isolated genomic DNA should be around 100 micrograms in quantity and have a purity of approximately 1.80 at the 260/280 absorbance ratio.2. DNA Fragment Generation
Once extracted, the DNA is broken into small, random-sized fragments using two techniques: restriction digestion and mechanical shearing. Restriction digestion involves using a restriction enzyme to cut the DNA into fragments. The enzyme binds to its specific recognition sites present multiple times throughout the genome, producing fragments with either sticky or blunt ends. Sticky ends are preferred as they are easier to insert into a vector. Alternatively, mechanical shearing produces blunt end cuts and requires an additional processing step for ligation into the vector.| NEET Biology Notes | ||
|---|---|---|
| Plasma Membrane Structure | Pre-Fertilisation | Economic Importance of Algae |
| Slime moulds | hypotonic solution | Post fertilisation |
3. Ligation of DNA into a Vector System
Next, the DNA fragments are inserted into a suitable vector. Common vectors include plasmids, cosmids, phages, bacteriophages, lambda phages, BACs, or YACs. The choice of vector depends on the size of the DNA fragment to be cloned. For example, a plasmid can carry up to 15kb of DNA, whereas a YAC system can accommodate fragments between 250-2000 kb. The vector is then processed to enable DNA fragment insertion, usually by digesting it with the same restriction enzyme used on the genomic DNA. The fragment and vector DNA are ligated using a ligase enzyme before being transformed into a host cell. It is also important to include a selectable or screenable marker, such as an antibiotic-resistant gene, to validate the insert.4. Transformation of Recombinant DNA into a Host Cell
In this step, the recombinant vector is inserted into a host cell, such as E. coli or yeast cells, where it replicates to produce copies of the genomic DNA fragments. The transformed cells are then cultured on agar media containing an antibiotic, ensuring that only cells with the antibiotic-resistant gene grow.5. Library Screening
Library screening identifies genes of interest using methods like colony hybridization and plaque hybridization, depending on the cloning vector used. Colony hybridization selects bacterial colonies containing the desired genes in plasmid- or cosmid-based libraries, while plaque hybridization identifies desired genes in phage-based libraries. Both methods use filter hybridization for primary screening of libraries through in situ replication on a nitrocellulose membrane.Genomic Library Uses
A genomic library is a collection of DNA fragments that represent an organism's entire genome. These fragments are stored in vectors and propagated in host cells. Here are the key uses: 1. Gene Identification and Cloning- Isolation of Genes: Helps in isolating and cloning specific genes for detailed study.
- Functional Analysis: Facilitates understanding of gene functions, expression patterns, and effects of mutations.
- Whole-Genome Sequencing: Essential for sequencing and assembling complete genomes.
- Comparative Genomics: Allows comparison of genomes from different species to understand evolutionary relationships and variations.
- Physical Mapping: Creates maps to locate genes and important genome regions.
- Linkage Mapping: Helps identify gene locations related to specific traits or diseases through inheritance patterns.
- Gene Expression Studies: Studies gene regulation and response under various conditions.
- Mutant Screens: Identifies genes involved in specific biological processes or diseases by screening for mutants.
- Disease Gene Discovery: Identifies genes associated with diseases by comparing healthy and diseased samples.
- Drug Target Identification: Discovers potential drug targets and therapeutic pathways.
- Gene Therapy : Develops gene therapies by identifying and cloning therapeutic genes.
- Crop Improvement: Identifies genes for desirable traits in plants, aiding in crop enhancement.
- Animal Breeding: Helps in identifying beneficial traits in animals for selective breeding programs.
- Phylogenetic Analysis: S tudies evolutionary relationships among species by comparing genomes.
- Genomic Diversity: Examines genetic diversity within and between species to understand adaptation and speciation processes.
Genomic Library and cDNA Library
The following table outlining the key differences between a Genomic Library and a cDNA Library:| Differences Between Genomic Library and a cDNA Library | ||
|---|---|---|
| Aspect | Genomic Library | cDNA Library |
| Source | DNA extracted from the entire genome of an organism. | mRNA extracted from a specific cell or tissue type. |
| Content | Contains all the genes and non-coding sequences of the genome. | Contains only the coding sequences (exons) of the expressed genes. |
| Purpose | To study the entire genome, including regulatory and non-coding regions. | To study gene expression and identify expressed genes in specific conditions. |
| Construction | Constructed by cutting genomic DNA into fragments and inserting them into vectors. | Constructed by reverse transcribing mRNA into cDNA and inserting it into vectors. |
| Applications | Genome mapping, functional studies, and cloning of genes and regulatory elements. | Gene expression analysis, identifying expressed genes, and studying alternative splicing. |
| Size of Inserts | Typically larger, ranging from 10 to 200 kb. | Typically smaller, usually representing only the coding regions (often less than 5 kb). |
| Library Complexity | Represents the entire genome, hence more complex. | Represents only the expressed portion of the genome, less complex. |
| Cloning Strategy | Uses restriction enzymes to cut DNA and ligate fragments into vectors. | Uses reverse transcriptase to synthesize cDNA from mRNA, which is then cloned into vectors. |
| NEET Biology Notes | |||
|---|---|---|---|
| Rhizopus | Fibrous root | Marchantia | Actinomycetes |
| Nostoc | Spirogyra | Ribosomes | Liverworts |
Genomic Library FAQs
Q 1: What is a genomic library?
Ans. genomic library is a collection of recombinant DNA molecules that includes the entire genome of an organism. It enables the storage and replication of overlapping DNA sequences, which can be analyzed to reconstruct the complete genome sequence using computational methods.
Q 2: What is the purpose of a gene library?
Ans. Gene libraries are utilized to discover new genes, sequence entire genomes, and compare genes across different organisms.
Q 3: What is a genomic library PDF?
Ans. genomic library is a set of clones sufficient in number to encompass all the DNA of a particular organism.
Q 4: What is a cDNA library?
Ans. cDNA library consists of cloned DNA molecules that are complementary to the mRNA of an organism. It represents a collection of cDNA, which are copies of the mRNA.
Q 5: How is a cDNA library constructed?
Ans. cDNA library is created by first binding complementary DNA (cDNA) to a vector, which is then used to transform bacterial cells. The transformed cells, containing plasmids with cDNA, are known as cDNA clones. This process often involves the use of a reverse transcriptase enzyme.
Q 6: What is a cloning vector?
Ans. Cloning vectors are DNA molecules that carry foreign DNA fragments when inserted. They can be bacterial plasmids, phasmids, cosmids, or bacteriophages, and must be capable of replicating independently.
🔥 Trending Blogs
Talk to a counsellorHave doubts? Our support team will be happy to assist you!

Check out these Related Articles
Free Learning Resources
PW Books
Notes (Class 10-12)
PW Study Materials
Notes (Class 6-9)
Ncert Solutions
Govt Exams
Class 6th to 12th Online Courses
Govt Job Exams Courses
UPSC Coaching
Defence Exam Coaching
Gate Exam Coaching
Other Exams
Know about Physics Wallah
Physics Wallah is an Indian edtech platform that provides accessible & comprehensive learning experiences to students from Class 6th to postgraduate level. We also provide extensive NCERT solutions, sample paper, NEET, JEE Mains, BITSAT previous year papers & more such resources to students. Physics Wallah also caters to over 3.5 million registered students and over 78 lakh+ Youtube subscribers with 4.8 rating on its app.
We Stand Out because
We provide students with intensive courses with India’s qualified & experienced faculties & mentors. PW strives to make the learning experience comprehensive and accessible for students of all sections of society. We believe in empowering every single student who couldn't dream of a good career in engineering and medical field earlier.
Our Key Focus Areas
Physics Wallah's main focus is to make the learning experience as economical as possible for all students. With our affordable courses like Lakshya, Udaan and Arjuna and many others, we have been able to provide a platform for lakhs of aspirants. From providing Chemistry, Maths, Physics formula to giving e-books of eminent authors like RD Sharma, RS Aggarwal and Lakhmir Singh, PW focuses on every single student's need for preparation.
What Makes Us Different
Physics Wallah strives to develop a comprehensive pedagogical structure for students, where they get a state-of-the-art learning experience with study material and resources. Apart from catering students preparing for JEE Mains and NEET, PW also provides study material for each state board like Uttar Pradesh, Bihar, and others
Copyright © 2026 Physicswallah Limited All rights reserved.








