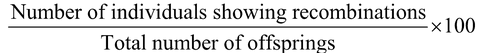Linkage
Chromosomal Basis of Inheritance of Class 12
Mendel’s law of inheritance are applicable to both chromosomes and genes. At the time of gamete formation, both maternal and paternal members of homologous chromosomes are independently distributed to gametes and hence the genes located on these chromosomes are also distributed independently.

Mendel’s law of independent assortment is not universally accepted and is applied only to genes present on different chromosomes and not to genes present on the same chromosome. An exception to Mendel’s law of independent assortment was reported by Bateson and Punnett (1906), who reported that the two pairs of alleles in Lathyrus (Sweet pea) do not show independent assortment. In first set experiments, they crossed two varieties of sweet pea, one with blue flowers and long pollens (BBLL) and other with red flowers and round pollens (bbll).
The results were :
- If there were independent assortment, the test cross ratio should have been 1 : 1 : 1 : 1, but here test cross ratio is found to be 7 : 1 : 1 : 7, i.e., parental combinations are in excess (Blue long, red-round). In this cross, the two dominants enter from the same parent and thus they try to remain together.
- In second set of experiments, Bateson and Punnett made a cross in which the two dominants enter from the different parents, i.e.,
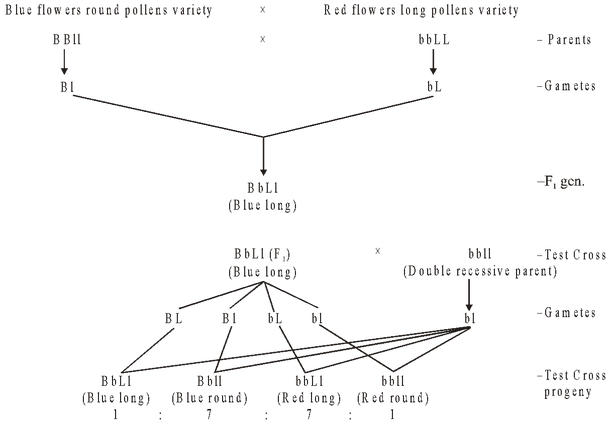
Again in this cross, test cross ratio comes out to be 1: 7 : 7 : 1 (not 1 : 1 : 1 : 1). In this case also, parental combinations are in excess (blue round and red long). So when the two dominants enter from the different parents, they try to remain separate.
Bateson and Punnett explained the above crosses as: When two dominants enter from the same parent, they try to remain together, which is called ‘coupling’.
On the other hand, when two dominants enter from different parents, they try to remain separate, which is known as ‘repulsion’.
T.H. Morgan (1910) reported the same thing in Drosophila and gave the term Linkage to this phenomenon. He also reported that coupling and repulsion are the two aspects of the linkage phenomenon.
Definition of linkage
“The tendency of parental combinations to remain together, which is expressed in terms of low frequency of recombinations (new combinations) is called linkage.”
Genes present on same chromosome thus show linkage. The parental combinations are due to linkage (new combinations or recombinations are due to crossing over).
According to T.H. Morgan : i.e.,
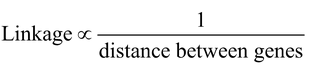
linkage is inversely proportional to the distance between the genes on chromosomes. Thus degree of linkage is dependent upon distance between linked genes on the chromosomes. More the distance between the genes, lesser will be the degree of strength of linkage and vice-versa.
Another example of linkage
It was reported by Hutchinson in Zea mays (maize). By crossing coloured and full endosperm variety with colourless shrunken variety and then by test crossing, he came to the conclusion that here again parental combinations are in excess (96.4%) which is due to linkage.
Significance of linkage
The possibility of variations (variability) in gametes is reduced by linkage (unless crossing over occurs). It provides a strong proof in favour of linear arrangement of genes on the chromosomes.
Linkage groups
All the genes linked together in a single chromosome (which do not show independent assortment), constitute a linkage group (Sutton, 1903). Number of linkage groups in an organism is equal to its haploid number of chromosomes, e.g., In Drosophila , n = 4, hence linkage groups = 4.
In Pisum sativum (garden pea), n = 7, hence linkage groups = 7. In Zea mays (maize), n = 10, hence linkage group = 10. Number of linkage groups in prokaryotes (bacteria, cyanobacteria or blue-green algae and mycoplasma) is one.
Crossing Over
According to T.H. Morgan, genes present on the same chromosome show linkage. Linkage may be complete (when only parental combinations are produced), e.g., in male Drosophila or incomplete (when some recombinations are also produced along with parental combinations). The recombinations or new combinations are inturn produced by a process, called crossing over. Thus crossing over is separation of linked genes (T.H. Morgan).
Definition of crossing over
“The process which produces recombinations or new combinations of genes as a result of interchange of corresponding segments between non-sister chromatids of homologous chromosomes.”
Crossing over occurs at pachytene sub-stage of prophase I of meiosis (at molecular level), but visible at diplotene sub-stage of prophase I. Chiasmata are the points, where crossing over occurs.
Crossing over distance between the linked genes, i.e., more the distance between genes on the chromosomes, more will be the chances of crossing over.
Crossing over may be single, double or multiple depending upon number of chiasmata which in turn depends upon length of the chromosome.
Crossing over is mostly two-stranded single crossing over with very few exceptions (although occurs at 4-strand stage in pachytene, but only 2 chromatids form chiasma where exchange occurs).
Germinal and somatic crossing over
- Germinal crossing over : Crossing over is common in germinal cells at the time of gamete formation (during meiosis) and it is called meiotic crossing over or germinal crossing over and is of great significance.
- Somatic crossing over : Generally pairing of homologous chromosomes occurs in meiosis in germinal cells, but sometimes this pairing of chromosomes also occurs in somatic cells and crossing over also occurs, which is called somatic crossing over.
This somatic crossing over has been reported in Drosophila by C. Stern, in maize by Jones and in Aspergillus nidulans (an ascomycetes fungus) by G. Pontecorvo.
Somatic crossing over occurs rarely (low frequency) and the mechanism is similar to germinal crossing over.
Example of crossing over
Crossing over was reported in Drosophila by T.H. Morgan in the following cross:
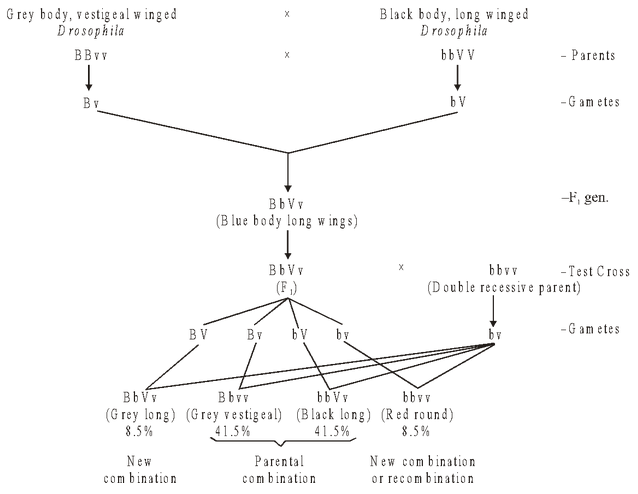
i.e. new combinations or recombinations have been reported in 17% cases, which are due to crossing over.
Theories regarding crossing over
Different theories of crossing over and chiasmata formation have been given as copy choice theory by Belling, breakage first theory by Muller, contact first theory by Serebrousky, etc. But the most accepted theory is precosity theory or strain theory, given by C.D. Darlington.
According to this theory, as a result of twisting of homologous chromosomes, a strain develops which causes breakage of chromatids at the point of contact. The broken segments unite with other segments to bring about crossing over, i.e., exchange of segments.
Factors affecting crossing over
- Temperature : Frequency of crossing over is increased by high and low temp.
- X-rays : Frequency of crossing over is increased by X-rays.
- Radium radiations : Radium radiations also increase crossing over.
- Sex : Crossing over is more in females.
- Age : Older females have more chances of crossing over.
Significance of crossing over
Crossing over provides a strong proof in favour of linear arrangement of genes on the chromosomes. Linkage maps or genetic maps are constructed on the basis of crossing over. Recombinations or new gene combinations are produced due to crossing over, which change genetic pool by changing of gene frequency and this provides a way of evolution (micro evolution).
Crossing over and linkage maps
As recombination frequencies or cross over frequencies are directly proportional to distances between genes, hence these values are used in formation of linkage maps (Sturtevant in Drosophila), i.e., relative distances between the genes. In linkage map, one map unit (m.u.) is considered equal to 1% recombination. A map unit is sometimes called centi Morgan (cM) after the name of Thomas Hunt Morgan. However, linear relationship between cM units and cross over frequencies does not hold good for long distances on a map. Recombination frequencies or cross over frequency or percentage of non-parental combinations can be calculated by the following formula: